Cancer Epigenetics
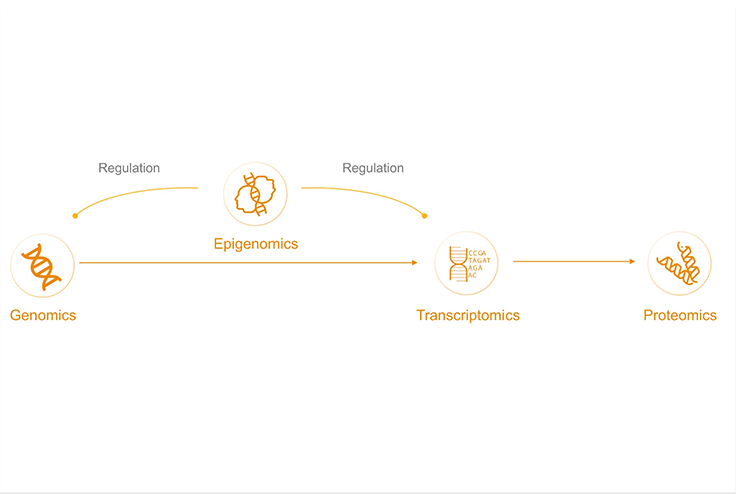
Why is epigenetics important to cancer research?
Cancer is a disease of the genome, but genetic mutations are only one factor. Non-genetic changes also affect the phenotype. Understanding the epigenetic landscape is increasingly crucial for modeling cancer initiation, progression, and therapeutic responses. Epigenomics looks at how cells control gene activity through processes such as DNA methylation.
Epigenomic technologies can identify cellular biomarkers associated with regulation of cancer genes or drug resistance:
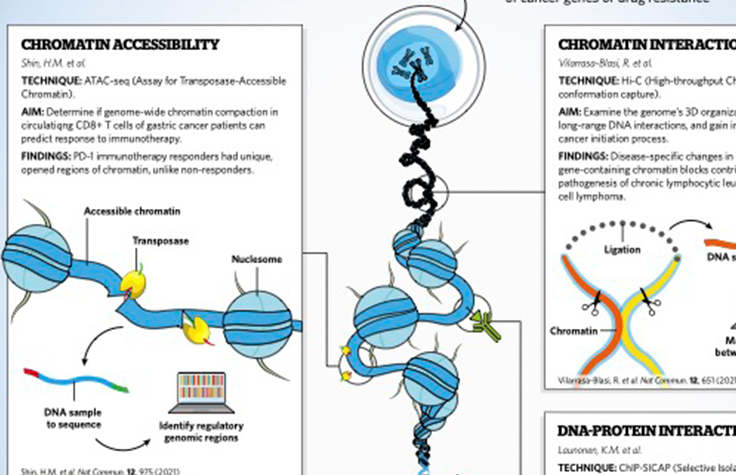
- Chromatin accessibility
- Chromatin interactions
- DNA modifications
- DNA-protein interactions
Methods to study epigenetic changes in cancer
ATAC-Seq
Cancer researchers can use ATAC-Seq to study epigenetic features across the genome without prior knowledge of regulatory elements. ATAC-Seq exposes genomic DNA to Tn5, a highly active transposase that preferentially inserts into open chromatin sites and adds sequencing primers. Subsequent NGS analysis, which includes genomic or transcriptomic profiling, provides insights into chromatin accessibility across the genome.
While several traditional methods like chromatin immunoprecipitation sequencing (ChIP-Seq), formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-Seq), or DNase I hypersensitive sites sequencing (DNase-Seq) can be used to study regions of chromatin–DNA interaction sites, ATAC-Seq illuminates regions of open chromatin.
Key advantages of ATAC-Seq
- Researchers can avoid sensitive enzymatic digestion of chromatin or rigorous validation of antibodies by using the Tn5 transposase
- It is possible to interrogate precious cancer samples with input requirements as low as 500-50,000 cells
- Researchers can obtain results rapidly, with a streamlined workflow that can be completed in < 3 hours
- ATAC-Seq can be paired with RNA-Seq to directly determine if regions of open chromatin are being expressed

Methylation arrays
Methylation arrays allow researchers to quantitatively interrogate methylation sites across the epigenome of cancer cells at single-nucleotide resolution. Array-based solutions provide comprehensive genome-wide coverage that includes but is not limited to CpG islands, CHH sites, enhancers, open chromatin, and transcription factor binding sites. As a high-throughput research method, the cost is minimal per sample compared with methylation sequencing alternatives.
Methylation array protocols have a user-friendly, streamlined workflow with >98% assay reproducibility and support for FFPE samples, increasing the applicability of methylation arrays to biobanked tissues.
Learn more about methylation arrays
Key advantages of methylation arrays
- Sample to data simplicity
- Compatibility with difficult sample types, such as FFPE samples
- Classification of tissue-specific differences between samples
- Low cost vs other methods
ChIP-Seq
Chromatin immunoprecipitation (ChIP) assays with sequencing (ChIP-Seq) is a powerful method to identify genome-wide DNA binding sites for transcription factors and other proteins. This method can reveal insights into gene regulatory events and biological pathways that play important roles in the development and progression of some cancers. Illumina offers efficient workflow solutions to enable you to perform genome-wide surveys of gene regulation using ChIP-Seq.
Key advantages of ChIP-Seq
- Captures DNA targets for transcription factors or histone modifications across the entire genome of any organism
- Defines transcription factor binding sites
- Reveals gene regulatory networks in combination with RNA sequencing and methylation analysis
- Offers compatibility with various input DNA samples
Learn more about epigenetics with our new guides
Cancer Research Methods Guide
An overview of Illumina sequencing followed by workflow suggestions for popular cancer research methods.

Multiomics Methods Guide
This guide provides multiomic research examples from recent literature along with detailed, end-to-end workflows. See recommendations for sample isolation, library prep, sequencing depth, data analysis, and more.
Epigenetic analysis products for cancer research
Infinium MethylationEPIC BeadChip Kit
- Microarray featuring expert-selected content
- Coverage of > 850,000 methylation sites per sample at single-nucleotide resolution
- FFPE compatibility
iScan Array Scanner
- High sample throughput
- Flexibility for multiple applications
- High data quality
- Automation compatibility
TruSeq ChIP Library Preparation Kit
- Generates chromatin immunoprecipitation sequencing (ChIP-Seq) libraries from ChIP-derived DNA
- Low DNA input requirement
- Multiplex sequencing with 24 unique indexes
Key cancer epigenetic analysis resources
Importance of epigenetics in cancer research
This infographic serves as a visual guide to see why epigenomics greatly impacts the understanding of cancer.

Understanding cancer biology with methylation microarrays
Cancer biology experts discuss the many advantages of leveraging methylation microarrays for biomedical research.

Advancing cancer research with multiomics
Learn how to link the causes and consequences of complex phenotypes through multiomics to enable discoveries that weren’t possible before.