“Whole-genome sequencing (WGS) is an emerging diagnostic approach that has already proved useful for the comprehensive identification of matching personalized targeted treatments and is now also demonstrating important potential for the identification of tumor tissue of origin, addressing a highly relevant unmet need for cancer patients for which the primary tumor is unknown or for cases where existing tumor (sub)type diagnostics is inconclusive.”
Edwin Cuppen, Director Hartwig Medical Foundation
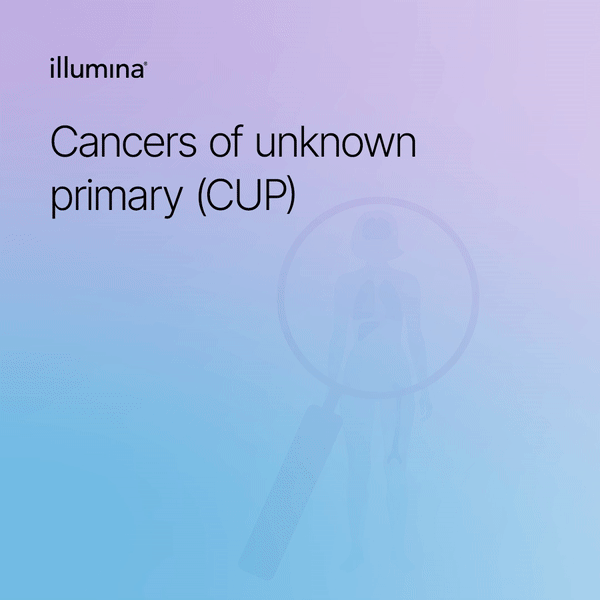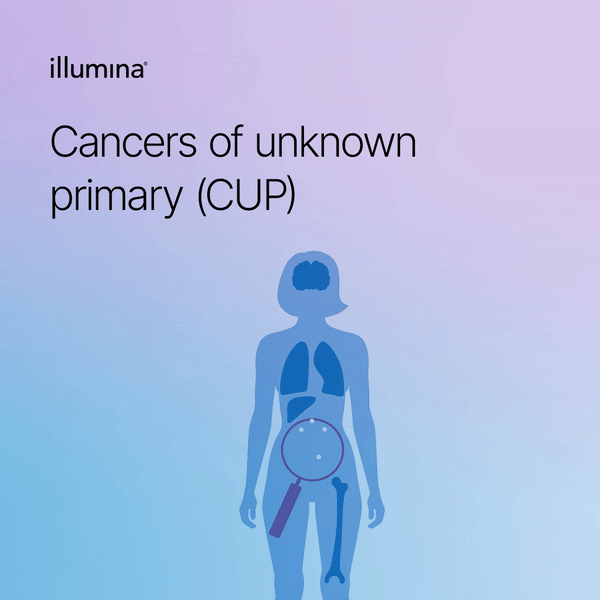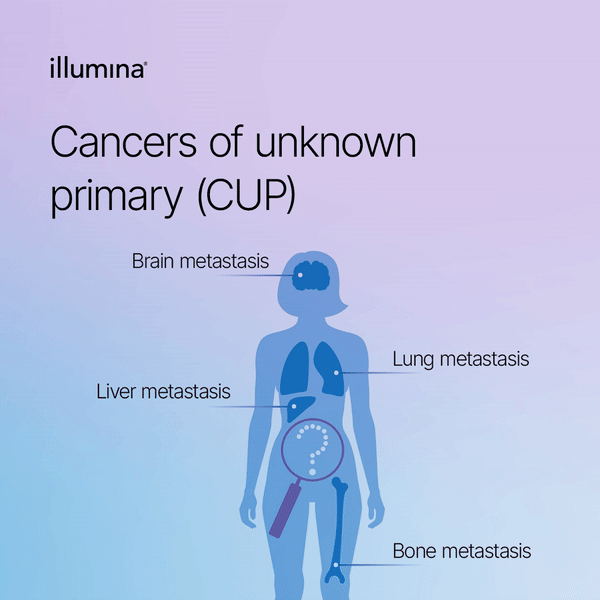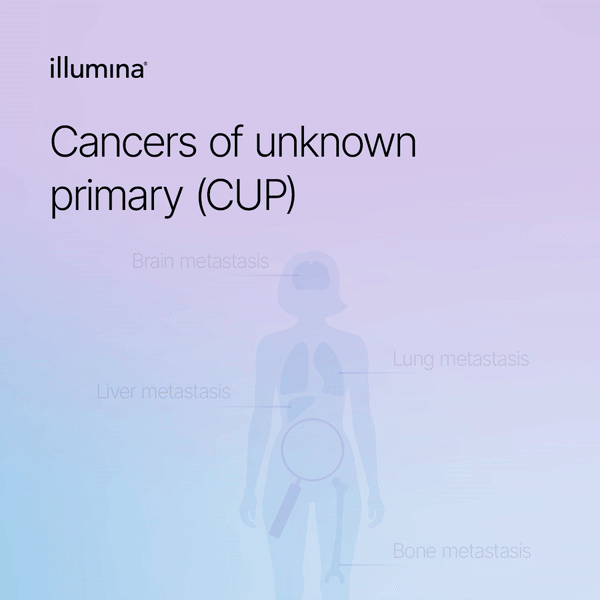 Figure 1. In carcinoma of unknown primary, cancer cells have spread in the body but the place where the primary cancer began is unknown.
Figure 1. In carcinoma of unknown primary, cancer cells have spread in the body but the place where the primary cancer began is unknown.
Cancers of unknown primary (CUP), also known as occult primary tumors, comprise a heterogenous group of metastatic neoplasms where the primary site of cancer cannot be determined, despite a thorough workup[1]. CUPs are rare, accounting for 2-5% of new cancer diagnoses worldwide, and have often a poor prognosis mainly due to the limited and non-selective treatment choices[2, 3]. Approximately 80% of CUPs do not respond to chemotherapy and have a median overall survival of <10 months[1]. The current diagnostic workup of CUP consists of extensive application of diagnostic approaches, including modern imaging, detailed histopathological and immunohistochemical investigations, and thorough physical and family history examinations.
Tissue of Origin (TOO) tests
The current standard method to diagnose tumor site of origin is immunohistochemistry (IHC), which has an accuracy of 60-70% for metastatic tumors[4]. When IHC is inconclusive, novel approaches to identify a likely tissue of origin (TOO) have been developed and utilize different technologies, including gene or miRNA expression analysis, DNA methylation, and next-generation sequencing (NGS). Gene expression profiling predicts the site of tumor origin based on differential gene expression patterns retained from the known primary tumor and have an accuracy ranging from 52.5-88.5% with varying numbers of tumor types discerned, depending on the assay[5, 6]. In addition, methods based on multi-omic approaches, including genomics and epigenomics, have also been applied to predict the TOO of CUP, claiming to increase the diagnostic range up to 30 different cancer types with up to 94% accuracy, especially when combined with machine learning and artificial intelligence[7,17]. Integration of NGS-based TOO tests into clinical practice has proven difficult due to hesitancy from the clinical community and the existing limitations of these tests, including sample requirements, accuracy, and lack of prospective studies. Moreover, the efficacy of site-specific treatment in patients with CUP remains controversial. Initial clinical studies have demonstrated mixed benefit of matching treatments to tumor types predicted by various TOO methods[5]. However, a recent meta-analysis showed that site-specific therapy might benefit patients with CUP predicted to originate from responsive tumor types[8].
Targeted therapies for CUP
The majority of clinical guidelines (e.g. European Society of Medical Oncology (ESMO) guidelines) now contain recommendations for the use of NGS to help identify tumor-specific genetic changes, which may be targeted with genotype-directed therapies[9]. In fact, CUP patients may be ideal candidates for personalized medicine, given the lack of registered therapies for CUP and growing number of tumor agnostic biomarker-directed targeted therapies and immunotherapies, like entrectinib or larotrectinib for NTRK-fusion positive tumors, or pembrolizumab for cancers with high tumor mutational burden (TMB) or microsatellite instability (MSI)[5]. The clinical impact of this approach has been evaluated in a small number of clinical trials, where treatment of CUP patients based on results of comprehensive genomic profiling (CGP) was compared with standard of care (SoC) approaches. The CUPISCO trial (NCT03498521) is an ongoing, phase II randomized study of targeted therapy or immunotherapy guided by CGP vs platinum-based chemotherapy in patients with CUP that have received 3 cycles of platinum doublet induction therapy. Retrospective analyses of CUP cases tested with CGP revealed 32% would match to a CUPISCO targeted therapy arm[10]. However, preliminary results from the trial revealed difficulties in screening patients due to complexities in CUP diagnosis, insufficient tissue for molecular analyses, and failure to meet inclusion criteria irrespective of CUP diagnosis[11]. The trial is ongoing and the primary outcome measures have yet to be disclosed. The CUP-COMP (NCT04750109) trial is an ongoing NHS initiative in the UK assessing tissue and liquid genomic sequencing for the diagnosis and treatment guidance in CUP patients, including a comparison of the effectiveness of tissue and blood-based biomarkers.
The value of Whole-Genome Sequencing
An increasing number of efforts evaluating the diagnostic and clinical utility of Whole-Genome Sequencing (WGS) are revealing potential applications where this sequencing method may bring improvements over existing diagnostic tests, providing not only mutational analysis, but also including the detection of homologous recombination deficiency (HRD), chromosomal instability (CIN), and other emerging genome-wide signatures that may guide tumor-specific treatments[12, 13]. For cancers of unknown primary, WGS offers a comprehensive DNA analysis that can identify diagnostic, prognostic, and predictive mutations, as well as a likely tissue of origin with a single test [14, 15]. Compared to targeted or whole-exome sequencing based TOO classifiers, WGS can detect more complex mutations, like structural variants, passenger events, and genome-wide mutation density distribution patterns that can be highly characteristic of tumor TOO and can increase the accuracy and classification of specific cancers[16]. Using features extracted from whole genome sequencing data of tumor-normal pairs and a machine learning approach, Nguyen and colleagues achieved an accuracy of ˜90%, discerning 35 cancer (sub)types[17]. The use of WGS for identifying clinically relevant genomic alterations has been shown in both retrospective and a limited number of prospective clinical studies, that have demonstrated that actionable genetic variants may be found in up to 71% of cancer patients[14, 18, 19]. The ongoing Dutch national prospective clinical trial, DRUP (Drug Rediscovery Protocol), aimed at describing the efficacy of matching therapy-refractory metastatic cancer patients to commercially available immuno or targeted therapies based on results of WGS, has also shown the ability of WGS to match a small cohort of CUP patients to targeted therapies[18]. As part of another Dutch study (WIDE), which was focused on implementation of WGS in routine diagnostic practice, more than 70 CUP patients from the Netherlands Cancer Institute were analyzed using WGS-based diagnostics by the Hartwig Medical Foundation, resulting in a definitive tissue of origin assignment in about 2/3 of all cases[20]. Furthermore, potentially actionable mutations were found in about 50% of patients. Follow-up data was available for a subset of 23 patients and 70% did start a treatment based on the WGS-based diagnostic outcomes, with about half driven by tumor type identification and the other half by the presence of a molecular biomarker.
From the patient perspective, the value of WGS cannot be minimized. WGS offers the possibility of not only reducing the endless tests and scans that are physically and emotionally taxing to patients, but also removes the uncertainty of non-specific treatments by identifying a potential TOO and/or targeted therapy options. Attending to this patient view along with recently published evidence on the diagnostic utility of WGS [21], in 2021, the Dutch Healthcare Authority (Nederlandse Zorgautoriteit, NZa) announced that WGS would be reimbursed for patients with metastasized cancer where the primary tumor cannot be located, and WGS is now taken up in the professional standard care protocol as the recommended diagnostic test to be performed for this cancer indication[20]. This approval may set precedent for the reimbursement of WGS for other cancer types, as well as for other countries looking to expand their personalized oncology programs. However, further evidence, including expanded data from prospective studies, of the ability of WGS to improve outcomes of CUP patients may be necessary for a wide implementation in routine clinical diagnostic testing. Meanwhile, the increasing adoption of WGS in routine diagnostics, should continue to forge a path for the development of WGS-based clinical applications.
Other Articles

Conference Spotlight: Prospective study supports use of WGTS in routine diagnostics of bone and soft tissue tumors.
From ESMO Sarcoma and Rare Cancers Congress 2023, an evaluation study by Karolinska Institute of Whole Genome and Transcriptome Sequencing (WGTS) for routine diagnosis of bone or soft tissue tumors.
References:
- Laprovitera N, Riefolo M, Ambrosini E, Klec C, Pichler M, Ferracin M: Cancer of Unknown Primary: Challenges and Progress in Clinical Management. Cancers (Basel) 2021, 13(3).
- Pavlidis N, Khaled H, Gaafar R: A mini review on cancer of unknown primary site: A clinical puzzle for the oncologists. J Adv Res 2015, 6(3):375-382.
- Pavlidis N, Pentheroudakis G: Cancer of unknown primary site. Lancet 2012, 379(9824):1428-1435.
- Anderson GG, Weiss LM: Determining tissue of origin for metastatic cancers: meta-analysis and literature review of immunohistochemistry performance. Appl Immunohistochem Mol Morphol 2010, 18(1):3-8.
- Kato S, Alsafar A, Walavalkar V, Hainsworth J, Kurzrock R: Cancer of Unknown Primary in the Molecular Era. Trends Cancer 2021, 7(5):465-477.
- Rassy E, Assi T, Pavlidis N: Exploring the biological hallmarks of cancer of unknown primary: where do we stand today? Br J Cancer 2020, 122(8):1124-1132.
- Abraham J, Heimberger AB, Marshall J, Heath E, Drabick J, Helmstetter A, Xiu J, Magee D, Stafford P, Nabhan C et al: Machine learning analysis using 77,044 genomic and transcriptomic profiles to accurately predict tumor type. Transl Oncol 2021, 14(3):101016.
- Ding Y, Jiang J, Xu J, Chen Y, Zheng Y, Jiang W, Mao C, Jiang H, Bao X, Shen Y et al: Site-specific therapy in cancers of unknown primary site: a systematic review and meta-analysis. ESMO Open 2022, 7(2):100407.
- Fizazi K, Greco FA, Pavlidis N, Daugaard G, Oien K, Pentheroudakis G, Committee EG: Cancers of unknown primary site: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2015, 26 Suppl 5:v133-138.
- Ross JS, Sokol ES, Moch H, Mileshkin L, Baciarello G, Losa F, Beringer A, Thomas M, Elvin JA, Ngo N et al: Comprehensive Genomic Profiling of Carcinoma of Unknown Primary Origin: Retrospective Molecular Classification Considering the CUPISCO Study Design. Oncologist 2021, 26(3):e394-e402.
- Pauli C, Bochtler T, Mileshkin L, Baciarello G, Losa F, Ross JS, Pentheroudakis G, Zarkavelis G, Yalcin S, Ozguroglu M et al: A Challenging Task: Identifying Patients with Cancer of Unknown Primary (CUP) According to ESMO Guidelines: The CUPISCO Trial Experience. Oncologist 2021, 26(5):e769-e779.
- Drews RM, Hernando B, Tarabichi M, Haase K, Lesluyes T, Smith PS, Morrill Gavarro L, Couturier DL, Liu L, Schneider M et al: A pan-cancer compendium of chromosomal instability. Nature 2022.
- de Luca XM, Newell F, Kazakoff SH, Hartel G, McCart Reed AE, Holmes O, Xu Q, Wood S, Leonard C, Pearson JV et al: Using whole-genome sequencing data to derive the homologous recombination deficiency scores. NPJ Breast Cancer 2020, 6:33.
- Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, Duyvesteyn K, Haidari S, van Hoeck A, Onstenk W et al: Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 2019, 575(7781):210-216.
- Roepman P, de Bruijn E, van Lieshout S, Schoenmaker L, Boelens MC, Dubbink HJ, Geurts-Giele WRR, Groenendijk FH, Huibers MMH, Kranendonk MEG et al: Clinical Validation of Whole Genome Sequencing for Cancer Diagnostics. J Mol Diagn 2021, 23(7):816-833.
- Jiao W, Atwal G, Polak P, Karlic R, Cuppen E, Subtypes PT, Clinical Translation Working G, Danyi A, de Ridder J, van Herpen C et al: A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat Commun 2020, 11(1):728.
- Nguyen L, Van Hoeck A, Cuppen E: Machine learning-based tissue of origin classification for cancer of unknown primary diagnostics using genome-wide mutation features. Nat Commun 2022, 13(1):4013.
- Hoes LR, van Berge Henegouwen JM, van der Wijngaart H, Zeverijn LJ, van der Velden DL, van de Haar J, Roepman P, de Leng WJ, Jansen AML, van Werkhoven E et al: Patients with Rare Cancers in the Drug Rediscovery Protocol (DRUP) Benefit from Genomics-Guided Treatment. Clin Cancer Res 2022, 28(7):1402-1411.
- Samsom KG, Schipper LJ, Roepman P, Bosch LJ, Lalezari F, Klompenhouwer EG, de Langen AJ, Buffart TE, Riethorst I, Schoenmaker L et al: Feasibility of whole-genome sequencing-based tumor diagnostics in routine pathology practice. J Pathol 2022.
- Schipper LJ, Samsom KG, Snaebjornsson P, Battaglia T, Bosch LJW, Lalezari F, Priestley P, Shale C, van den Broek AJ, Jacobs N et al: Complete genomic characterization in patients with cancer of unknown primary origin in routine diagnostics. ESMO Open 2022, 7(6):100611.
- Samsom KG, Bosch LJW, Schipper LJ, Roepman P, de Bruijn E, Hoes LR, Riethorst I, Schoenmaker L, van der Kolk LE, Retel VP et al: Study protocol: Whole genome sequencing Implementation in standard Diagnostics for Every cancer patient (WIDE). BMC Med Genomics 2020, 13(1):169.

 Upgrade your browser or select an alternate
Upgrade your browser or select an alternate Chrome
Chrome Firefox
Firefox Safari
Safari Edge
Edge