Introduction
Pharmacogenomics (PGx) can empower research into drug efficacy, appropriate dosage, and genetic risks for adverse drug reactions. By taking into account the status of genetic factors known to impact drug metabolism in each genome, researchers can better understand how existing drugs behave and increase the chances of gathering enough statistical evidence to support new drug development.
For technical reasons, many of these genes can be challenging to assess. Laboratories need tools for measuring copy number status, resolving interference from homologous genomic regions, and combining individual calls into merged gene-level results called “star alleles.” For the first time, all these tools are now available to researchers and laboratories performing PGx analysis using whole-genome sequencing (WGS).
In this post, we describe a WGS-based integrated PGx star allele caller for the 20 most critical PGx genes we embedded into Illumina’s DRAGEN 4.0 Bio-IT platform. We will cover how the various methods work and demonstrate the pipeline’s effectiveness on both gold-standard and real-world data sets.
Glossary of commonly used terms
The purpose of PGx is to predict drug metabolism, drug response, and potential adverse drug reactions. Collectively, these traits are called “phenotypes.” The phenotype can be predicted based on the “genotype," that is, the combination of “haplotypes.” Haplotypes are sets of genetic variants present on the same chromosome and can include single nucleotide polymorphisms, insertion/deletion, and/or copy number variants. Since many PGx haplotype designations contain an asterisk (as in *1/*2), haplotypes are often called “star alleles.” Because we all have two copies of most PGx genes, the final piece of the puzzle is to combine the two detected haplotypes into pairs, called “diplotypes.”
Gene and variant selection
To support consistency across the field of PGx testing, the Clinical Pharmacogenetics Implementation Consortium (CPIC)1 curates and posts freely available, peer-reviewed gene/drug clinical practice guidelines. As of March 2022, CPIC classifies 20 genes as having strong evidence for gene-drug interactions (“Level A”). These are the genes that should always be included when performing PGx analysis. Other databases, such as Pharmacogenomic KnowledgeBase (PharmGKB),2 Pharmacogene Variation (PharmVar),3 and the Food and Drug Administration (FDA) “Section 1” list4 provide additional details.
We used these resources to inform which genes and variants to include in our caller, focusing first on the 20 genes marked by CPIC as “Level A.”
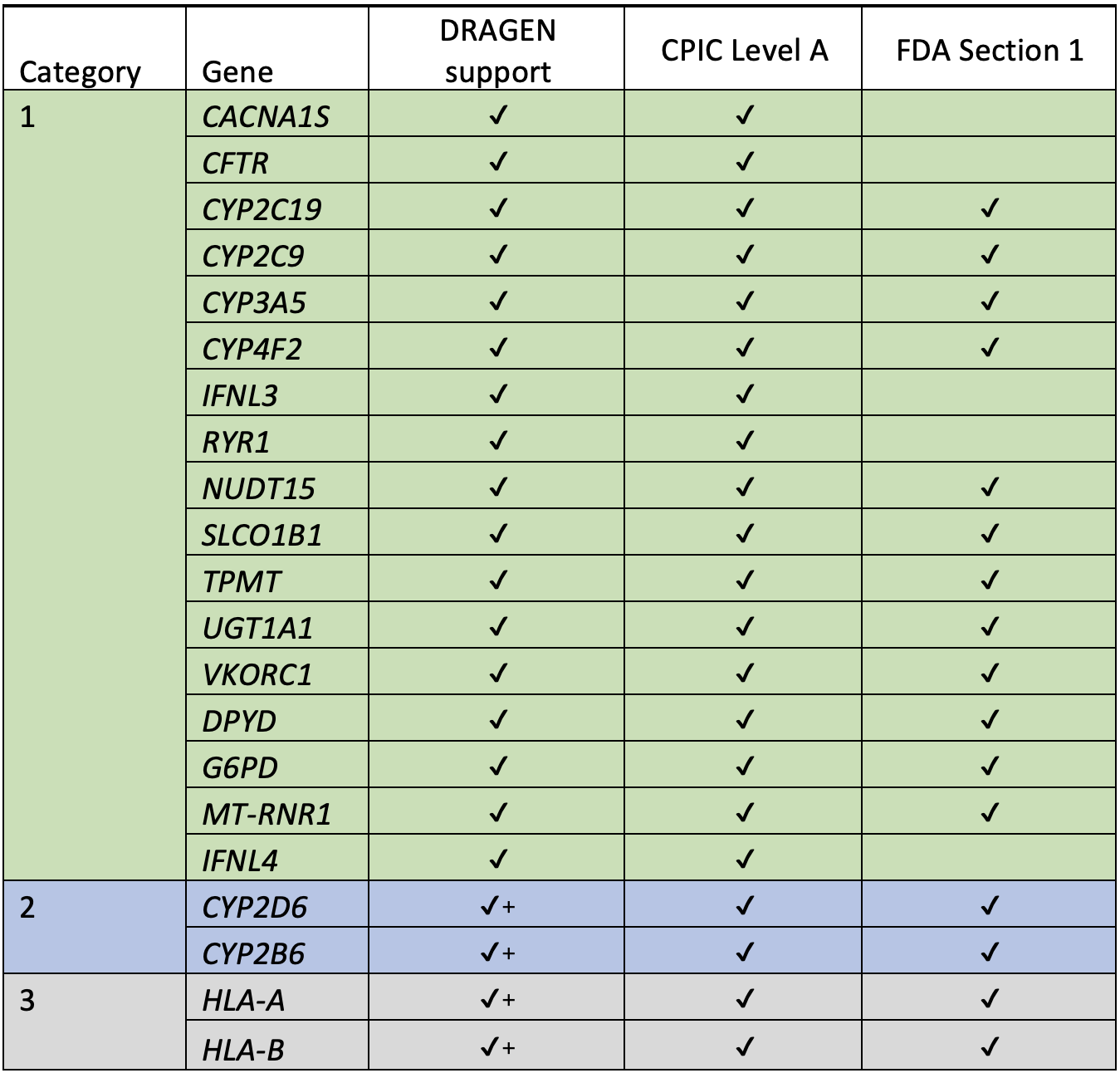
For the “DRAGEN support” column:
✓ = support for this gene in DRAGEN 4.0+ for GRCh38
✓+ = support for this gene in DRAGEN 4.0 for both GRCh38 and hg19
Approaches to genotyping
Some genes are more challenging than others to genotype with next-generation sequencing technologies. Variants from 16 of 20 genes currently supported in DRAGEN v4.0 can be called with high accuracy using the standard DNA sequencing mapping and variant calling pipeline; however, four genes cannot and require special attention: CYP2B6 and CYP2D6 due to paralogous regions in the genome, and HLA-A and HLA-B due to high levels of polymorphism.
Thus, we divide the list of 20 genes currently supported in DRAGEN v4.0 into three method categories:
PGx category 1: Standard variant calling
The first category solved by the star allele caller genotypes the following 16 genes using standard mapping and variant calling, then uses predefined haplotype combinations from PharmGKB to compose the most likely diplotype. These are the genes in category 1:
CACNA1S, CFTR, CYP2C19, CYP2C9, CYP3A5, CYP4F2, IFNL3, RYR1, NUDT15, SLCO1B1, TPMT, UGT1A1, VKORC1, DPYD, G6PD, MT-RNR1
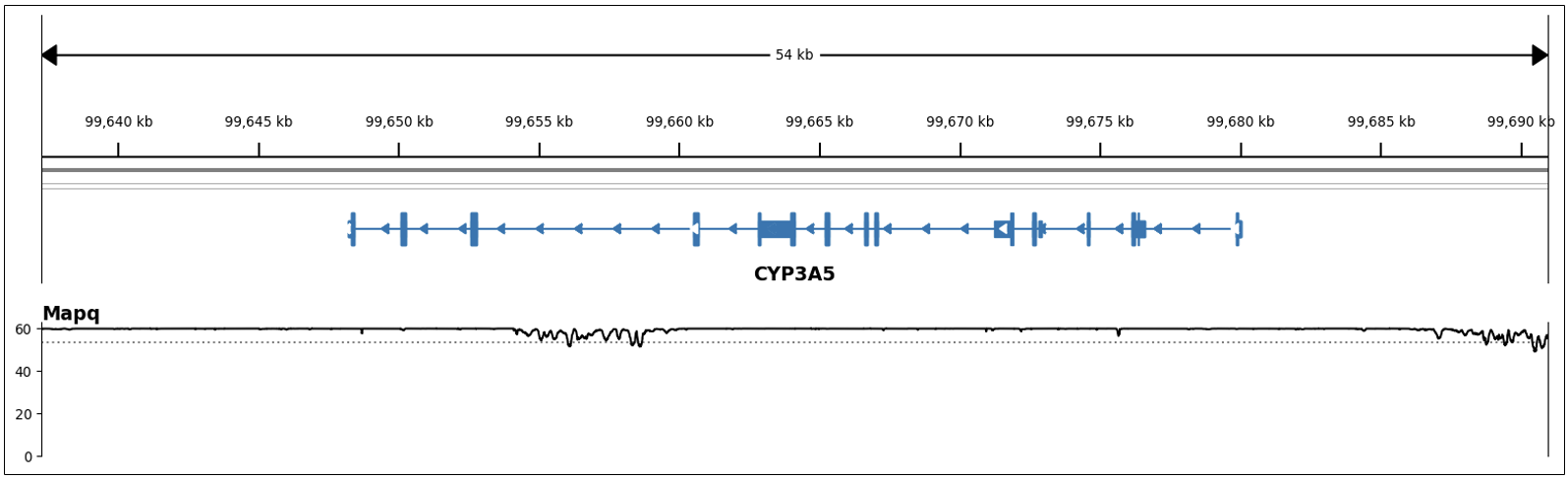
Variants within these genes lie on easy-to-map regions of the genome; that is, these genes do not share high homology with other parts of the genome. Hence, reads align accurately and uniquely to these genes, which enables accurate variant calls. For instance, in Figure 1 we can see the high and consistent MAPQ across the span of CYP3A5, a star allele caller gene.
In addition to outputting the star allele for each gene, the caller also includes the metabolism status in the PGx report, by directly using a genotype-metabolism status mapping resource from PharmCAT.5
PGx category 2: Targeted paralog callers
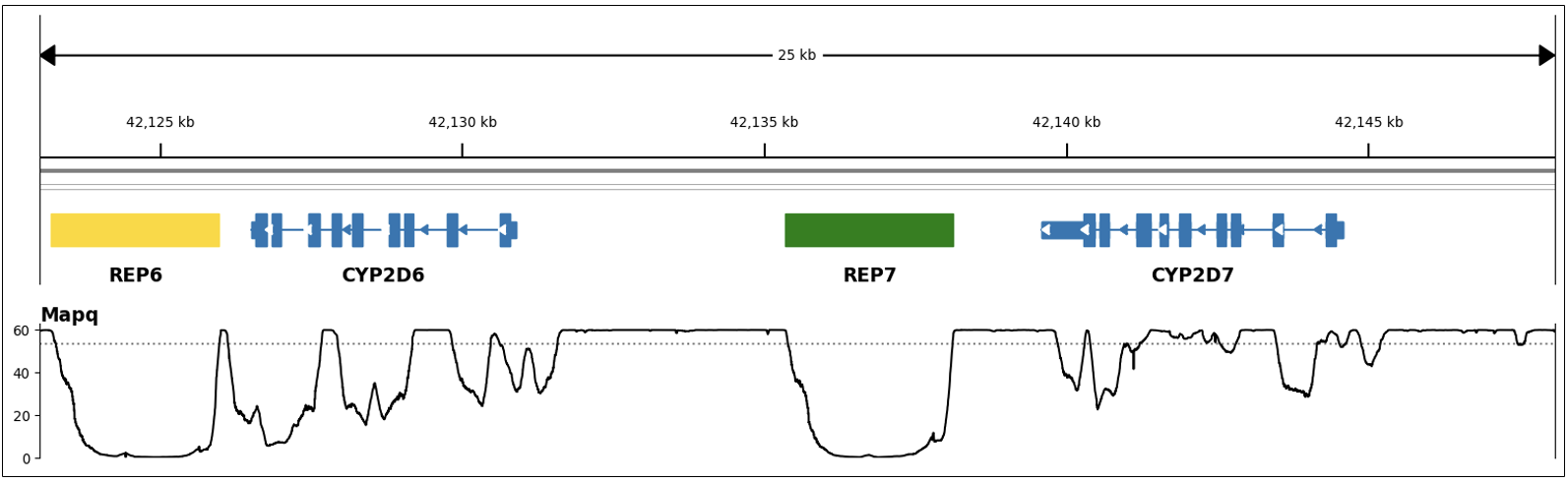
The second category contains genes CYP2D6 and CYP2B6, which are called by special purpose targeted callers.6 Genotyping these genes is challenging through standard variant calling approaches due to the presence of paralogous pseudogenes. CYP2B6 has one pseudogene (CYP2B7) with ~93% shared sequence homology and CYP2D6 has two pseudogenes: CYP2D7 (~97% homology) and CYP2D8P (~91% homology). This homology reduces read mapping accuracy for these genes (as illustrated in Figure 2, showing regions with low MAPQ) and makes variant calling error prone. Additionally, these genes are characterized by a large number and variety of common structural variants not identifiable through standard small variant calling.
The targeted callers solve this problem by combining copy number analysis, structural variant calling, and small variant calling on targeted regions of the genome where the genes are located. Variant phasing is performed using reads aligned to differentiating sites between the gene and pseudogene(s). Star allele annotations for called variants are extracted from PharmVar and the caller also adds the metabolism status in the report, using the mapping resource from PharmCAT.5
PGx category 3: Targeted HLA callers
The third category deals with genotyping HLA-A and HLA-B genes, which are highly polymorphic; that is, these genes have thousands of defined haplotypes across the population, each containing hundreds of variants. These genes are genotyped by another special purpose workflow called the HLA caller. This caller uses expectation maximization to analyze reads aligning to full sequence alleles from the IMGT/HLA database7 and Allele Frequency Net Database8 to output two-field resolution of HLA.
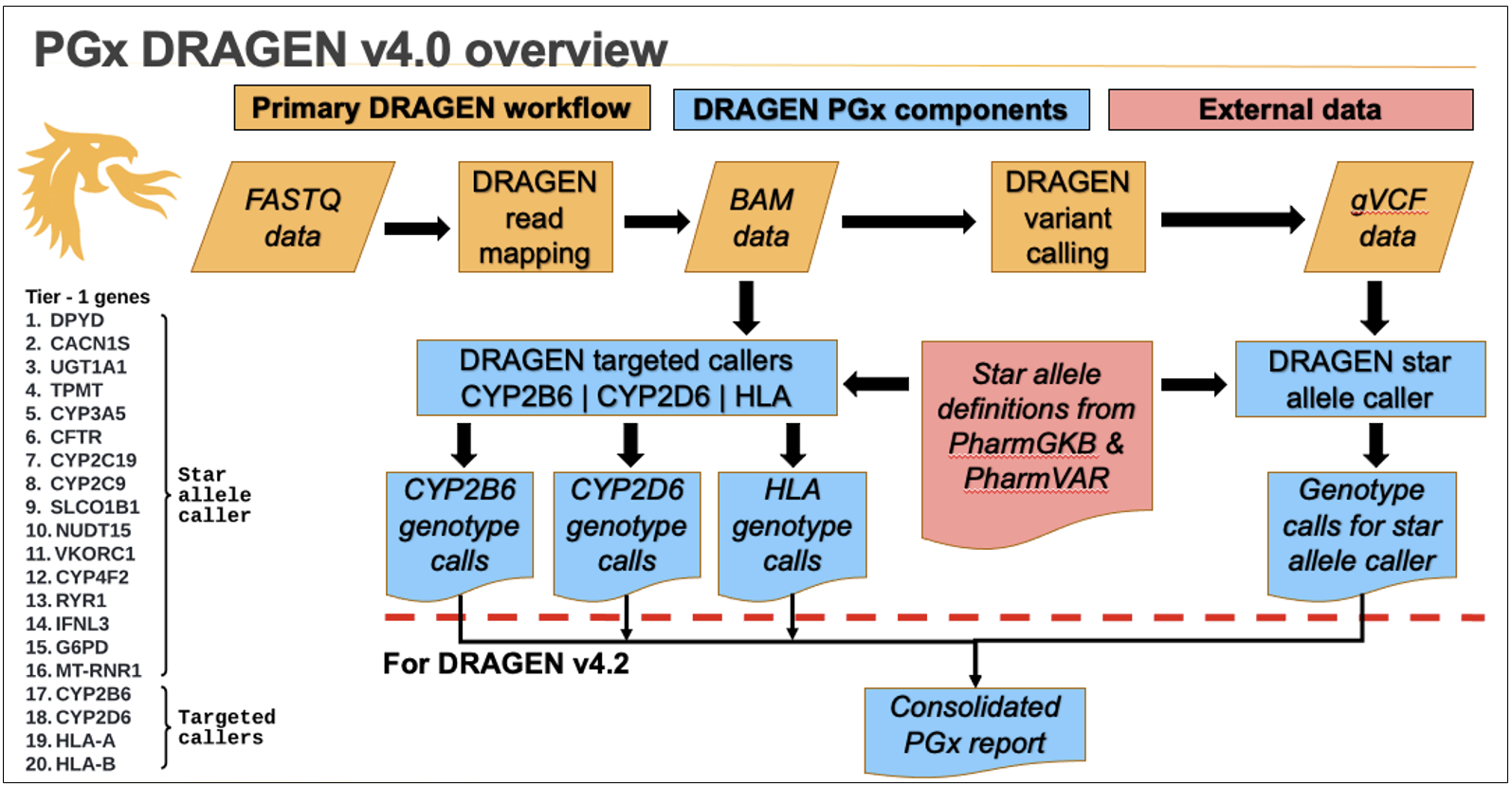
DRAGEN PGx workflow
The overall DRAGEN PGx workflow is described in Figure 3. In DRAGEN v4.0, the star allele caller and targeted callers each output their own file separately.
Validation of callers
Internal validation of category 1 genes
The DRAGEN v4.0 star allele caller was comprehensively tested for 15 out of the 16 genes with available data by comparing its output against external PGx callers (PharmCAT and Aldy9) for cell lines. We found 100% concordance across these genes (Table 2).
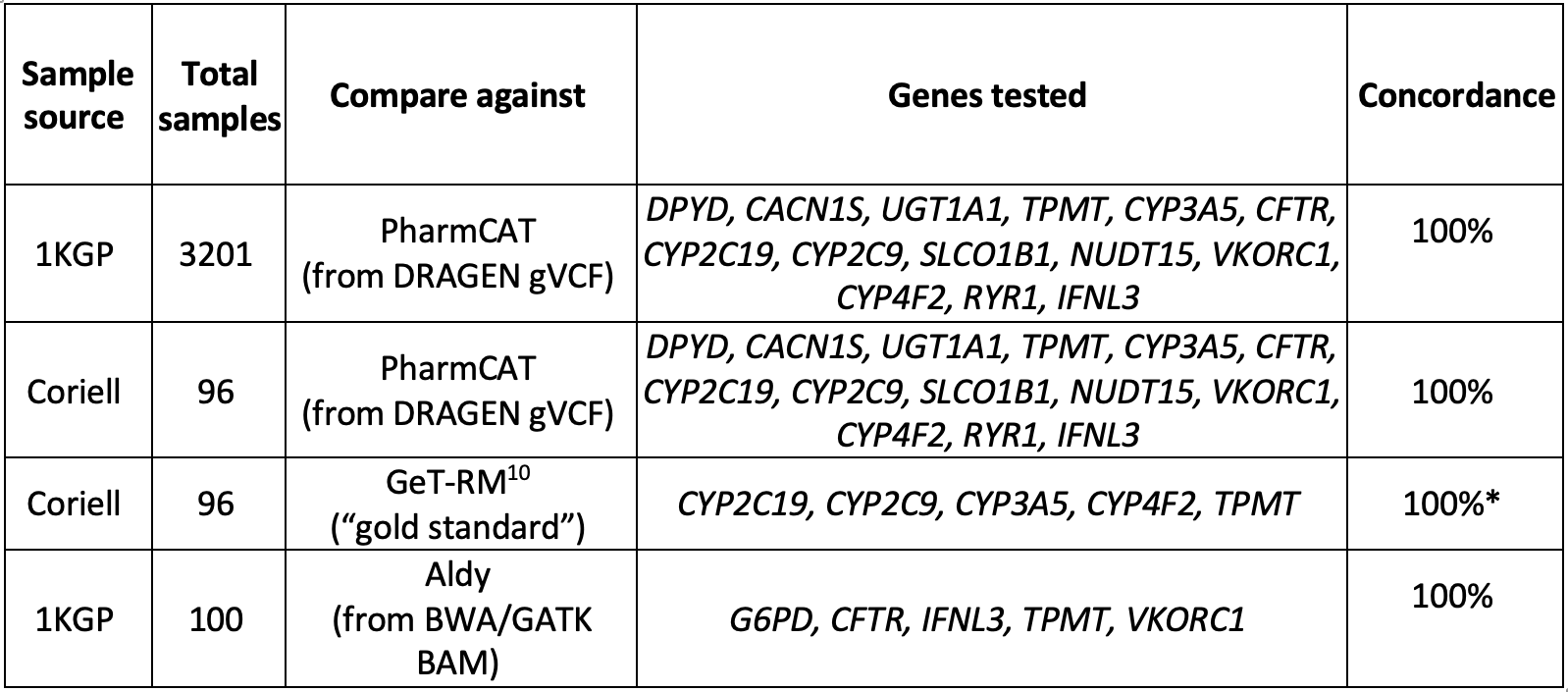
*Some calls were excluded from our analysis because the definitions for one or more of their alleles have changed since their publication.
External validation of category 1 genes
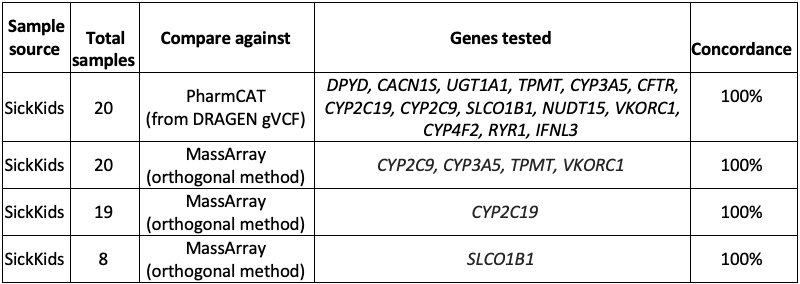
External collaborators at the Hospital for Sick Children (SickKids11) in Toronto conducted an independent validation of the DRAGEN v4.0 star allele caller. They performed orthogonal bioinformatics (PharmCAT) and wet lab testing (MassArray12) on 14 genes over 20 samples.
The analysis found 100% concordance between DRAGEN v4.0 star alleles and both PharmCAT and MassArray for the various combinations of samples and genes tested (Table 3).
CYP2D6 caller
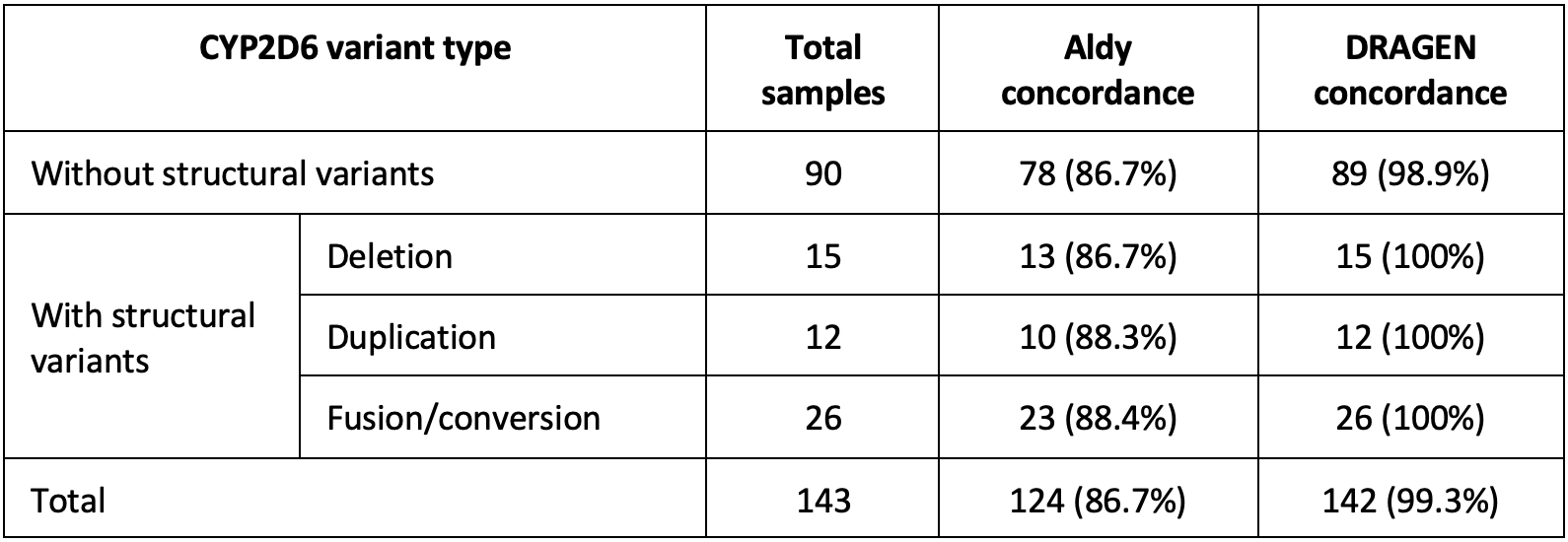
The CYP2D6 caller was validated against 143 variant samples from Coriell13 and the 1000 Genomes Project (1KGP).14 Calls were compared against either GeT-RM or manually curated calls from long-read sequencing data. Shown in Table 4 below are the concordance results of the DRAGEN v4.0 CYP2D6 caller along with the concordance of Aldy using the same input sequencing data. Concordance against the reference call-set is higher for DRAGEN than for Aldy.
CYP2B6 caller
The DRAGEN CYP2B6 caller was validated against 125 samples from 1KGP and 76 Coriell samples. Calls on the Coriell samples were compared against either GeT-RM or calls from Stargazer.15 For the six samples where calls were not concordant with GeT-RM, DRAGEN was concordant with Stargazer on five samples and Stargazer did not make a call on the sixth sample. Calls on the 125 samples from 1KGP were compared against calls manually curated from long-read sequencing data. Shown in Table 5 below are the concordance results of the DRAGEN v4.0 CYP2B6 caller.
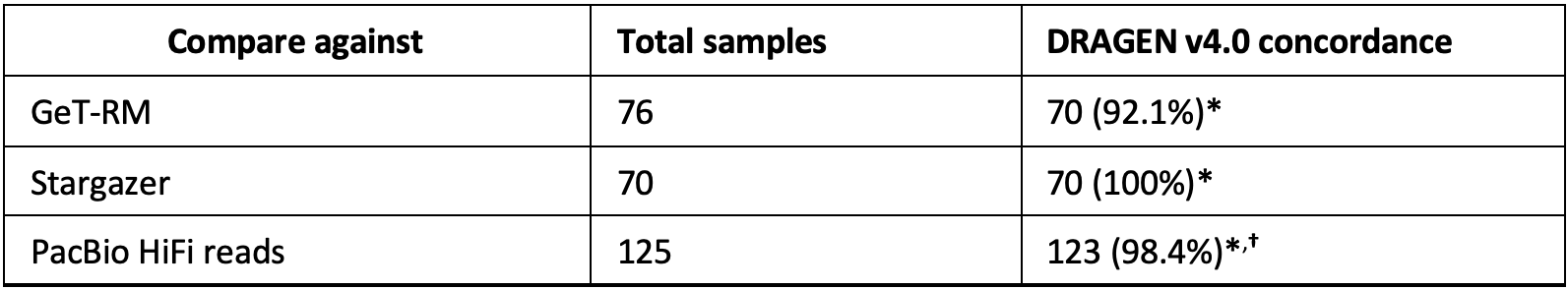
*Concordance based on metabolizer status when multiple genotypes are reported (for example, *1/*6;*4/*9).
†Two discordant samples have a novel star allele (*U1) resulting in multiple possible genotypes,
HLA caller
The DRAGEN v4.0 HLA caller was validated for genotyping accuracy with 117 WGS samples from the 1000 Genomes consortium.14 Of the 351 calls (three genes for each of the samples), 349 calls were concordant with Sanger sequencing (accuracy 99.6%).
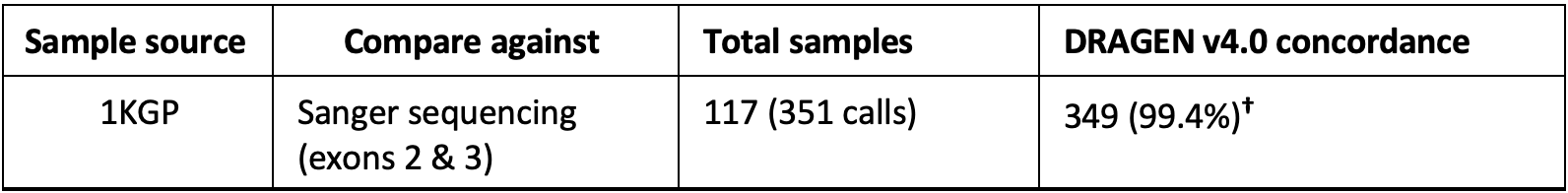
†The overall accuracy shown is an aggregate across the three HLA genes in DRAGEN v4.0: HLA-A, HLA-B, and HLA-C.
Future directions
Because PGx is a relatively new field of study, our understanding of which genes and variants impact PGx outcomes will continue to evolve. We’ll watch out for any developments and do our best to make sure DRAGEN is keeping up.
During the software development cycle for DRAGEN 4.0, CPIC added ABCG2 to the list of Level A genes. Several other genes are strong candidates to be included in future releases, including NAT2, BCHE, and UGT2B17.
In the next DRAGEN release (4.2), we plan to expand support to the hg19 human genome reference, though only a subset of the category 1 genes have star alleles defined in the hg19 reference.
Additional post-processing is being planned to merge all PGx results into a single output file (Figure 3).
For more information or a DRAGEN trial license for academic use, please contact dragen-info@illumina.com.
References
1. Clinical Pharmacogenetics Implementation Consortium
2. PharmGKB
3. PharmVAR
4. FDA PGx recommendations
5. PharmCAT
6. Cyrius
7. IMGT/HLA DB
8. AFND
9. ALDY
10. GeT-RM PGx study
11. SickKids
12. MassArray
13. Coriell
14. 1000 Genomes Project
15. StarGazer