Gene Expression Analysis
Reveal mechanisms of cell activity through gene expression analysis
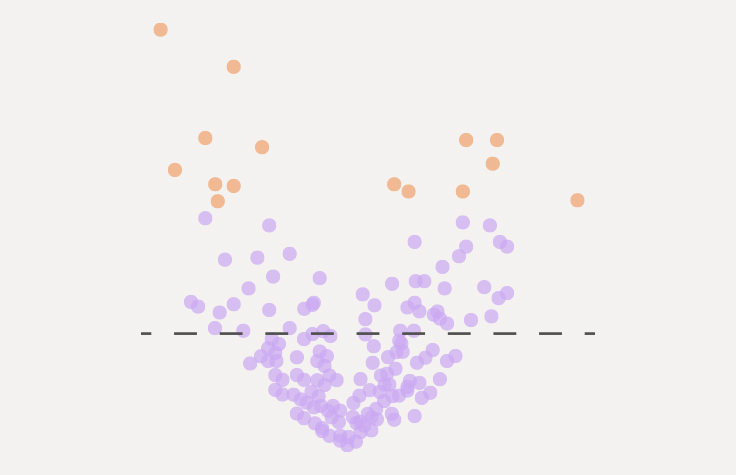
What is gene expression analysis?
Gene expression analysis examines transcriptional gene expression of the coding region of the genome. This method allows sensitive and accurate quantification of gene expression by analyzing mRNA, identification of known and novel isoforms in the coding transcriptome, detection of gene fusions, and measurement of allele-specific expression.
Advanced technologies such as next-generation sequencing (NGS) have revolutionized the scale, speed, and accuracy of profiling gene expression levels in a single experiment. Compare NGS-based RNA sequencing (RNA-Seq) to other common gene expression profiling methods, such as gene expression microarrays and qRT-PCR, and learn how to analyze gene expression and identify novel transcripts using RNA-Seq.
Benefits of gene expression profiling with RNA-Seq
Explore the advantages of NGS for analysis of gene expression, gene regulation, and methylation.
Download eBookCommon gene expression analysis methods
A variety of methods may be used to profile gene expression for select targets of interest and/or analyze the coding transcriptome, based on your study goals. Learn about the benefits and considerations of several key gene expression and transcriptome analysis methods.
| Method | Benefits | Considerations |
|---|---|---|
RNA-SeqLearn more: |
Can detect both known and novel features in a single assay. Broad dynamic range. Can be applied to any species. Scalable and cost-effective for high-throughput applications. |
Sequencing low numbers of targets (e.g. less than 10 targets) can be time-consuming. |
Gene expression microarraysLearn more: |
High sample throughput for analysis of known genes and transcripts. Familiar workflow for many scientists. |
Inability to detect novel transcripts. Gene expression measurement is limited by background at the low end and signal saturation at the high end.1 |
qRT-PCRLearn more: |
Effective for low target numbers (e.g., less than 10 targets). Quick, simple workflow using equipment already in most labs. |
Can only detect known sequences. Low scalability. |

Workflows for RNA sequencing
Explore our comprehensive guide detailing Illumina solutions for next-generation RNA sequencing applications.
Download GuideAdditonal resources
Gene expression patterns in breast cancer
In episode 41 of the Illumina Genomics Podcast, Dr. Ake Borg discusses classification of breast tumors based on patterns of gene expression, methylation, and other genomic alterations.
Searching for gene expression profiles associated with cancer
Researchers discuss how they use RNA-Seq and other NGS methods to uncover cancer-associated gene expression biomarkers.
Uncovering drug-susceptible tumorigenic pathways
Researchers use NGS-based RNA-Seq to profile biomarkers and analyze transcriptomic signatures of activated pathways in cancer samples.
Cancer gene expression profiling
Analyzing gene expression and transcriptome changes with RNA sequencing can help researchers understand tumor classification and progression.
Measuring gene expression from single cells
Highly sensitive ultra-low-input and single-cell RNA-Seq methods enable researchers to explore the biology of individual cells in complex tissues and understand cellular subpopulation responses to environmental cues.
Complex disease research
Gene expression and transcriptome profiling studies can help researchers better understand neurological, immunological, and other complex diseases on a molecular level.
References
- Zhao S, Fung-Leung WP, Bittner A, and Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;16;9(1):e78644.