Illumina next-generation sequencing (NGS) has been the most widely adopted technology for human whole-genome sequencing (WGS), delivering an accurate, scalable, cost-effective solution, featured in over 300,000 scientific publications.1 However, a small portion of the genome remains challenging to map due to highly repetitive or highly homologous regions.
Illumina Complete Long-Read technology, previously announced as 'Infinity', will address these edge cases and accelerate access to the remaining ~5% of genic regions that are challenging to map. Illumina long-read technology uses a proprietary library prep leveraging trusted Illumina SBS chemistry, the accuracy and speed of DRAGEN analysis, and the scalability of Illumina Connected Analytics to achieve high-performance long-read data.
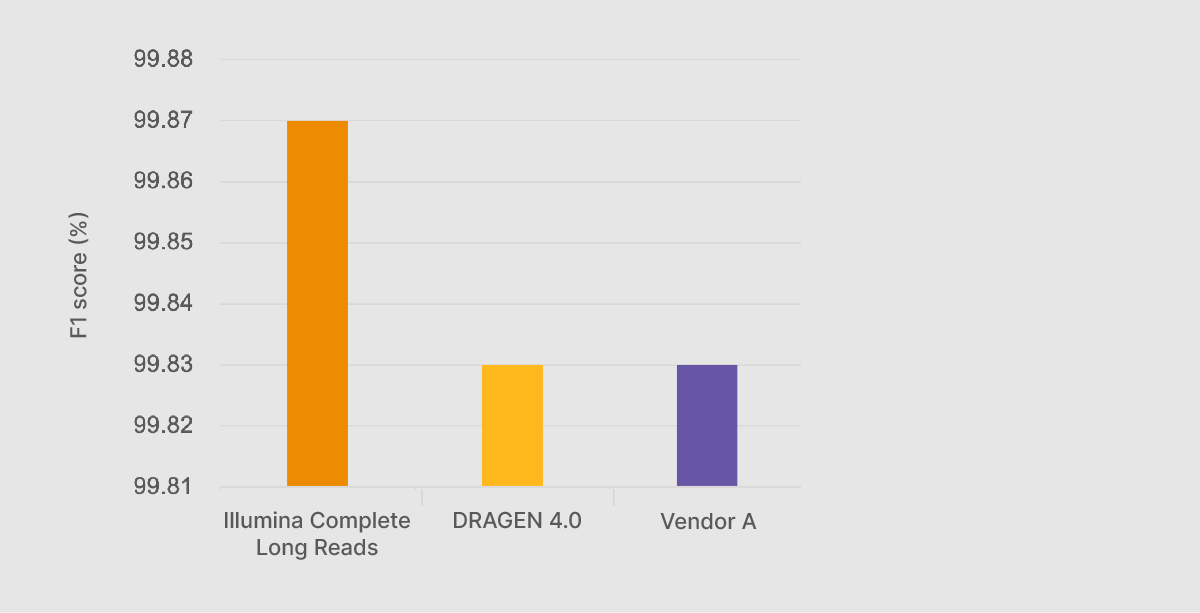
At the Illumina Genomics Forum in September 2022, Chief Science Officer Alex Aravanis presented preliminary Illumina Complete Long-Read performance data against the benchmarking data sets from the PrecisionFDA Truth Challenge v2.2 Illumina Complete Long Reads with DRAGEN analysis generated an F1 score—a compound statistic of precision and recall—of 99.87%, higher than any other method, including on-market long-read technologies (Figure 1).
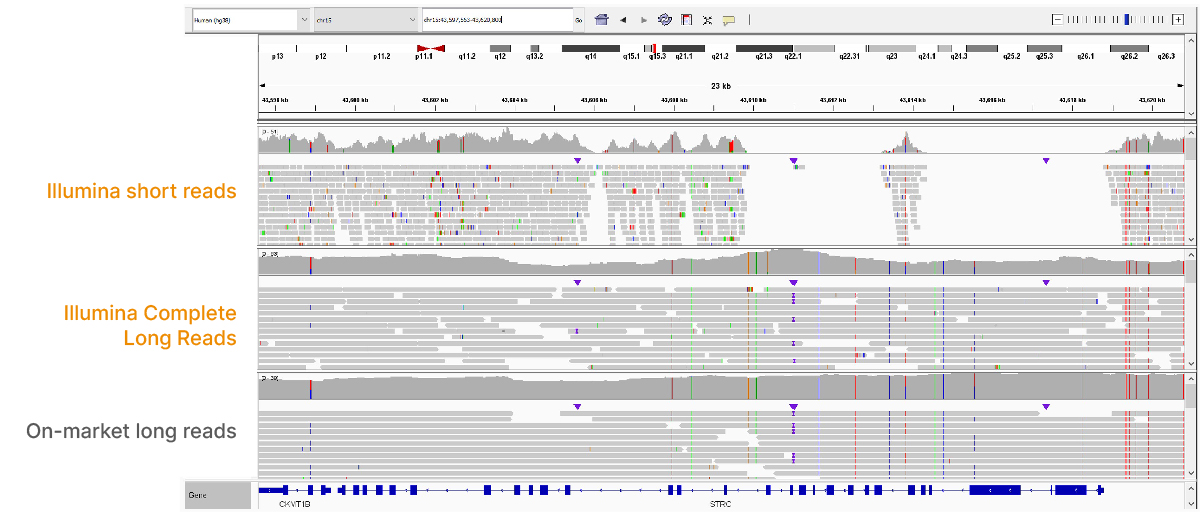
The Illumina Complete Long-Read assay demonstrates improved performance, accessibility, and scale relative to current on-market long-read solutions. The assay uses a standard NGS workflow to generate contiguous long-read data with N50 of 6–7 kb, including read lengths > 30 kb for human WGS (Figure 2, Figure 3). The efficient, single-day library preparation makes it easy to scale for high-throughput studies. The protocol is also compatible with many sample types, requiring only 50 ng DNA input with no specialized extractions, shearing, or size selection.
Multiple products based on the Illumina Complete Long-Read assay are in development:
- Illumina Complete Long-Read Prep, Human (launching in Q1 2023) designed for human WGS
- Illumina Complete Long-Read Prep with Enrichment for human WGS, with targeted long-read data focused on the most challenging genic regions

Demonstrated power of the Illumina Complete Long-Read assay
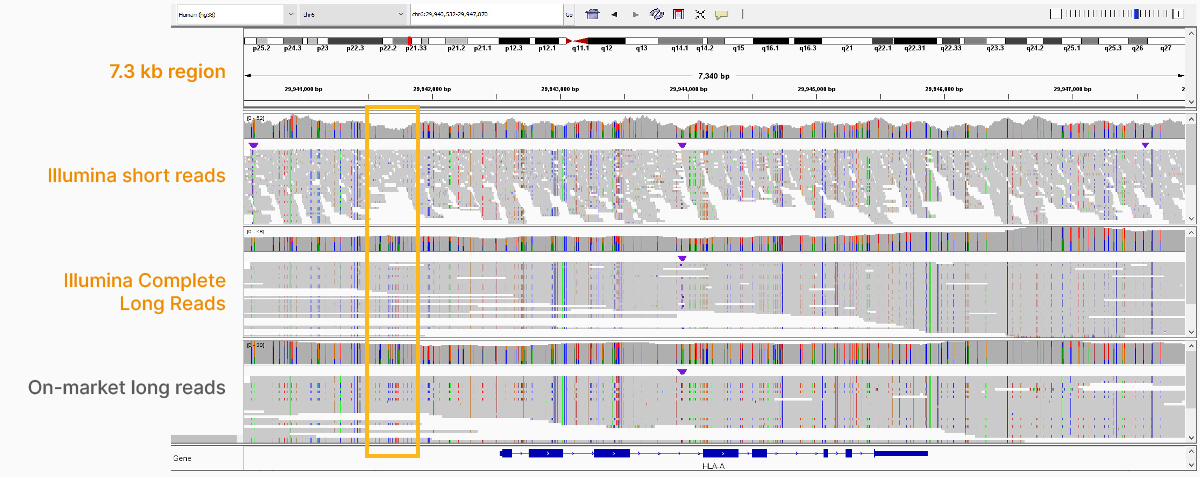
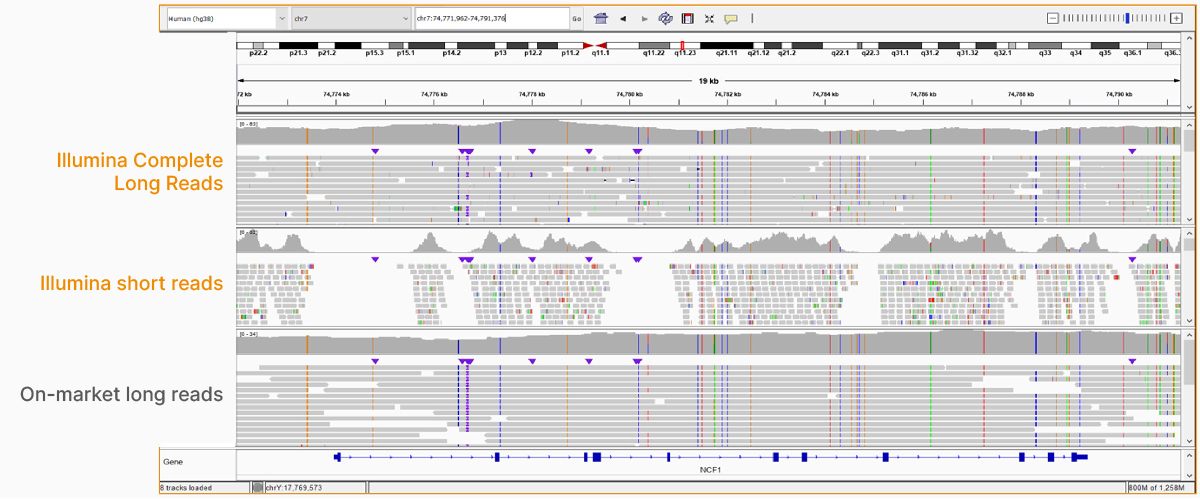
At an April 2022 webinar, we demonstrated the ability of Illumina long-read data to improve alignment and variant calling in traditionally challenging regions like repetitive regions, highly polymorphic regions (Figure 4), pseudogenes and paralogs (Figure 5), large insertion–deletion variants (indels) (Figure 6), and structural variants (Figure 7).
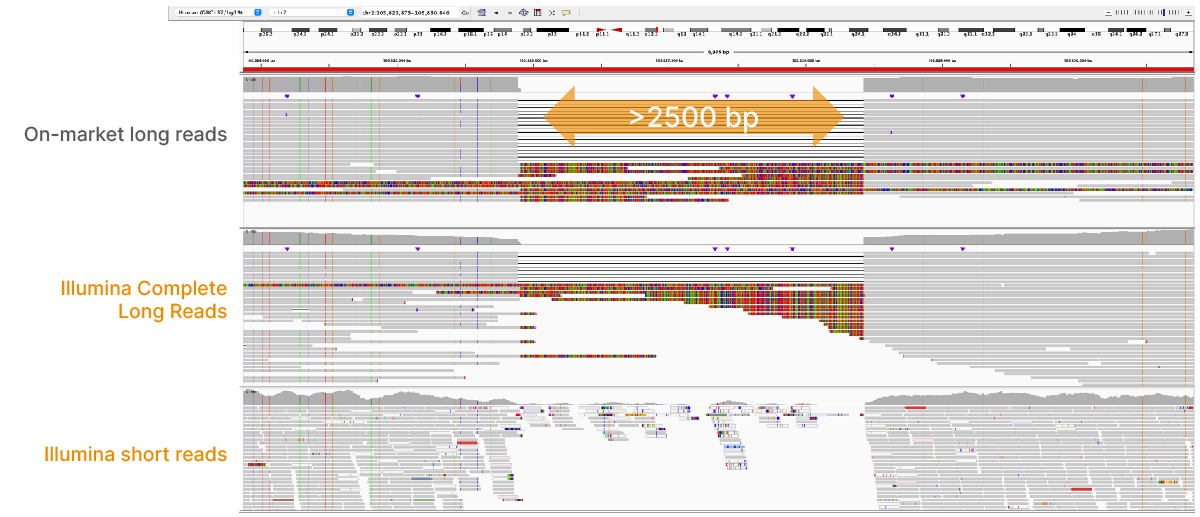
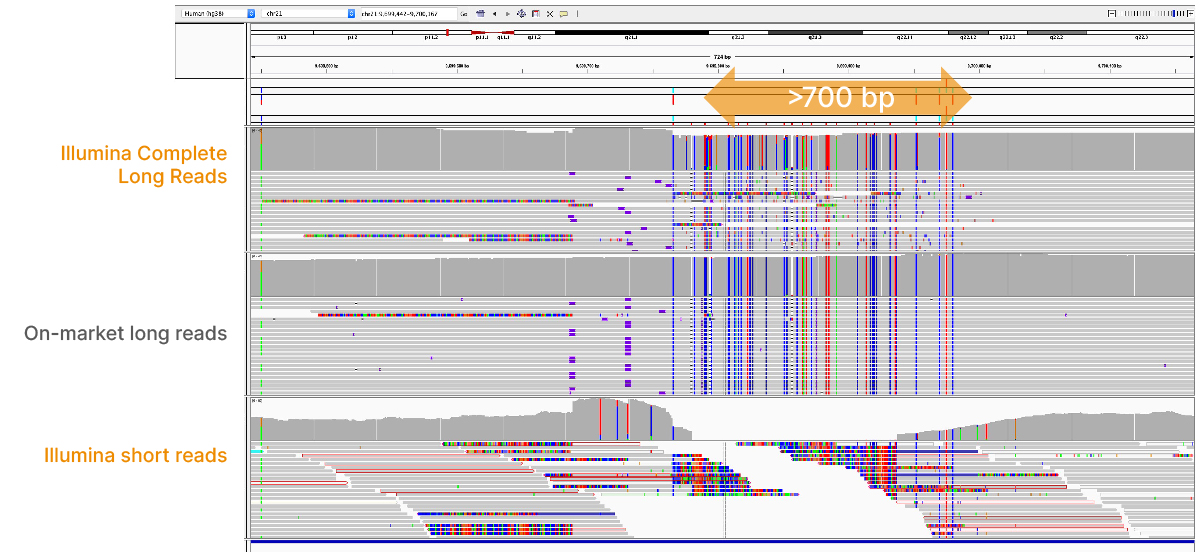
In addition to the whole-genome assay, we showed how the Illumina Complete Long-Read technology is compatible with enrichment methods (Figure 8). Targeted solutions focus on regions known to benefit from additional insight with longer reads. Future products with enrichment can create additional flexibility and scalability.

Illumina Complete Long-Read technology is advancing our understanding of the human genome
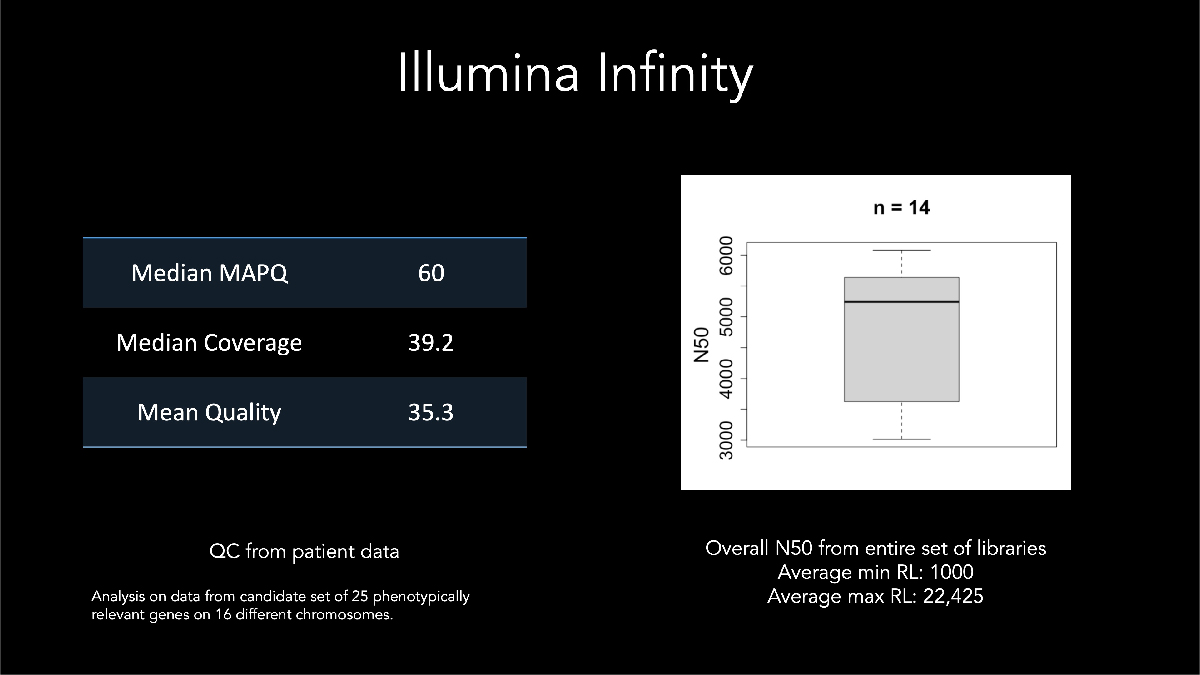
In his keynote address at AGBT 2022, Dr Euan Ashley of Stanford Medical Center described using Illumina long-read data for calling and phasing a de novo variant with a sample from a patient affected by genetic disease. Dr Ashley highlighted early development assay metrics where samples generated median N50 > 5 kb with some reads exceeding 22 kb (Figure 9).
At the 2022 Illumina Genomics Forum, the n-Lorem Foundation presented an update regarding their collaboration with Illumina to sequence n-Lorem patient samples using the Illumina Complete Long-Read assay. The resulting phased high-quality sequencing enables the identification of patient-specific SNPs associated with the pathogenic allele that are essential for patient-specific design of antisense oligonucleotides. These data enable n-Lorem to discover personalized antisense oligonucleotide (ASO) medicines for patients with genetically defined nano-rare diseases (fewer than 30 individuals globally).
Tracy Cole, PhD, Sr. Director of Research at n-Lorem said, "The accuracy and cost-effectiveness of the Illumina Infinity technology are important to us at n-Lorem, but even more important to our patients who are in desperate need of help. The ability to accurately call rare variants and phase data into haplotypes helps inform our ASO discovery process and ensures that we have the correct data to make important drug discovery decisions."
Illumina remains committed to extending the breadth of applications we support and delivering the most complete and comprehensive view of the genome.
References
- Data calculations on file, Illumina, Inc. 2022.
- PrecisionFDA Truth Challenge V2. precision.fda.gov/challenges/10. Published 2020. Accessed September 20, 2022.