Spatial transcriptomics of complex tissues
Learn about high-resolution, high-throughput spatial transcriptomics of kidney tissues using the NanoString GeoMx Digital Spatial Profiler and Illumina sequencing systems.
Spatially resolve transcriptional activity in complex tissue architectures using RNA-Seq

Spatial transcriptomics combines molecular profiling with spatial context to show where genes are active within intact tissue. Techniques such as immunohistochemistry (IHC) and in situ hybridization localize specific targets within tissue architecture, while newer methods extend this capability to transcriptome-wide analysis. By integrating RNA sequencing (RNA-Seq) powered by next-generation sequencing (NGS) with spatial analysis, researchers can access a previously unavailable view of the transcriptome within its morphological context.
A key benefit of spatial transcriptomics is the ability to map gene expression across tissue sections, linking molecular activity with tissue structure. This spatial resolution enables researchers to study cellular interactions in complex tissues such as tumor microenvironments, compare normal and diseased regions, and build quantitative atlases of tissue function.
While imaging-based methods like IHC and in situ hybridization reveal specific markers, RNA sequencing-based spatial transcriptomics extends this capability to thousands of genes simultaneously, delivering a comprehensive molecular profile within preserved tissue architecture. With NGS-based spatial transcriptomics, researchers can:
Access resources to help plan and implement spatial transcriptomics studies. Explore workflow options, data analysis tools, and integration capabilities.
Your email address is never shared with third parties.
Imaging-based approaches typically use fluorescent in situ hybridization to visualize transcripts directly within intact tissue, achieving subcellular resolution across selected gene panels. These methods reveal fine spatial patterns of gene expression with exceptional precision.
Sequencing-based approaches capture transcripts from defined tissue regions, preserving spatial information through spatially encoded capture areas. After NGS, reads are mapped back to their original locations to reconstruct spatial gene expression patterns, enabling cell-level resolution across large tissue regions.
These approaches allow researchers to link molecular activity with tissue structure and reveal spatial relationships underlying biological function.
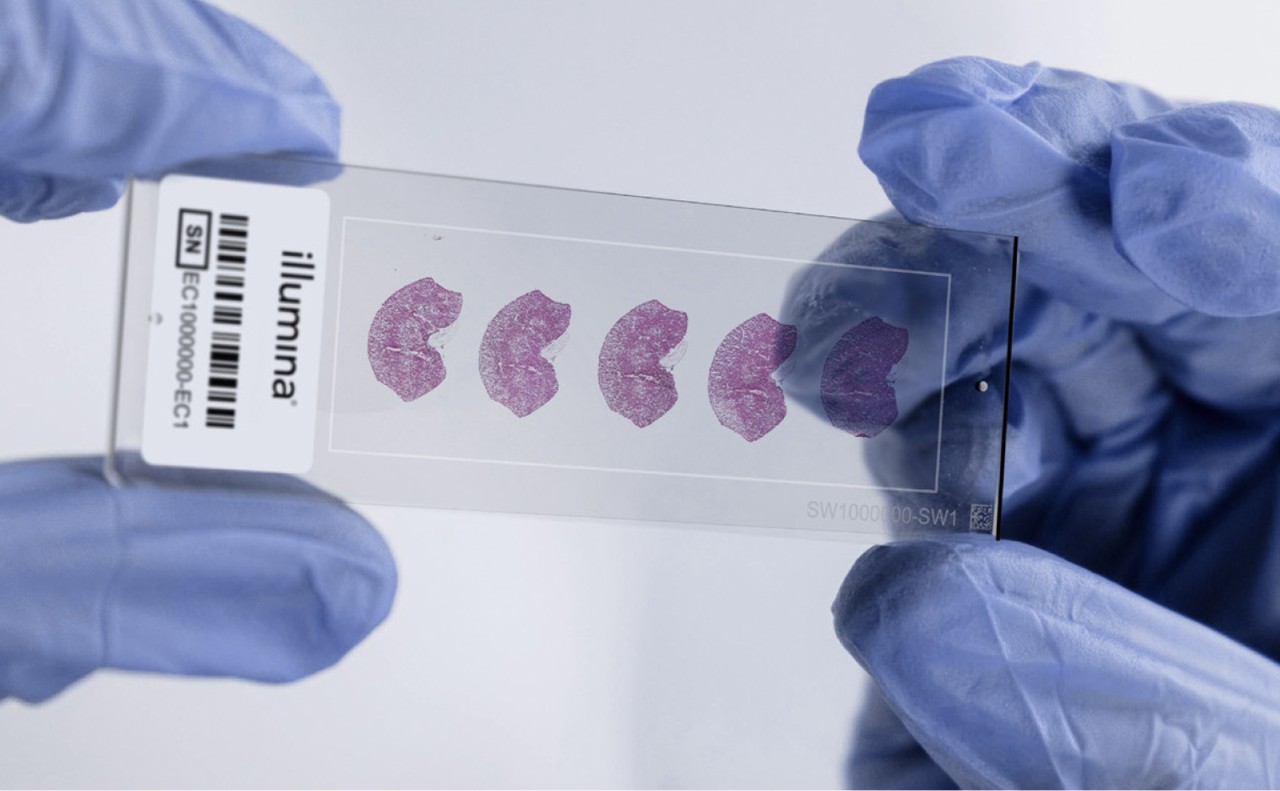
Tissue preparation for spatial transcriptomics
Tissue sections are mounted, fixed, stained, and imaged to provide histological context. These preparatory steps preserve tissue morphology and enable the subsequent generation of spatially resolved gene expression data.
Imaging-based methods capture subcellular detail, revealing fine spatial patterns within tissue architecture. Sequencing-based approaches combine imaging with NGS to achieve cell-level resolution and whole-transcriptome coverage across large tissue areas. Sequencing can provide comprehensive gene expression insights without compromising resolution or coverage breadth.
Illumina spatial technology offers a high-resolution, sequencing-based approach for whole-transcriptome analysis in intact tissue. It provides broad transcriptomic coverage across large tissue areas, a key advantage over targeted or imaging-based methods.
Spatial biology can incorporate other analytical methods such as epigenomic, genomic, and proteomic information at the cellular level while similarly providing contextual information within preserved tissues. These methods collectively fall under the term spatial multiomics and offer a multidimensional approach to comprehensively understand biological systems. For many spatial multiomic applications, Illumina offers NGS-powered technologies that can link structural, functional, and spatial insights to enable your next discoveries.

Download our in-depth eBook on how multiomic methods have revolutionized research through cutting-edge sequencing and array technologies.
Your email address is never shared with third parties.
Analyze whole transcriptomes in FFPE tissue sections in a spatially resolved manner by mapping gene expression over a high-resolution microscope image.
Characterize hundreds to thousands of RNAs in cells and tissues with subcellular resolution using in situ imaging.
Profile expression of RNA and protein from distinct tissue compartments and cell populations with an automated, scalable workflow.
Perform high-resolution in situ imaging for spatial transcriptomics across whole tissue samples.
Spatial transcriptomics of complex tissues
Learn about high-resolution, high-throughput spatial transcriptomics of kidney tissues using the NanoString GeoMx Digital Spatial Profiler and Illumina sequencing systems.
Transcriptional profiling of tissue sections
Leverage Visium Spatial Gene Expression from 10x Genomics for transcriptional profiling of entire tissue sections.
Single-cell and spatial sequencing on NextSeq 1000 and 2000 Systems
Learn how XLEAP-SBS chemistry combined with 10x Genomics single-cell and spatial solutions enable high-resolution genomics on the NextSeq 1000 and NextSeq 2000 Systems.
Spatial proteomic and proteogenomic insights
Combine the spatial analyses of RNA and protein using the Nanostring GeoMx DSP and Illumina sequencing to understand the heterogeneous pathology of human astrocytoma and glioblastoma.
Advances in spatial biology and cancer research
Scientific experts trace the evolution of cancer research using solutions from NanoString and Illumina from a spatial biology perspective.
Spatiotemporal changes in cardiac tissue
Learn about transcriptional changes across cardiac tissue regions during early-onset acute myocardial infarction.
Single-cell transcriptomic landscape in Alzheimer’s disease
See how Dr Vivek Swarup’s application of spatial transcriptomics has led to advancements in understanding late-stage Alzheimer’s disease.
Learn how spatial multiomic profiling can provide unprecedented molecular information in cancer biopsy samples.
Read how spatial information is providing next-generation improvements in multiomic data for cancer research.
Read how investigators use spatial transcriptomics to create an atlas of the developing human immune system.
Learn how researchers use spatial transcriptomics to identify differences in ovarian tumor microenvironments, gene expression, and therapeutic responses.
See how scientists discovered the spatial layout of diverse cells in various tissues using spatial transcriptomics.
Read how Dr Alex Swarbrick uses single-cell techniques to study tumor microenvironments in breast and prostate cancers.
Single-cell RNA sequencing can provide solutions to unravel complex biological systems at high resolution with minimal sample input.
Introduction to transcriptomics
See how a comprehensive view of transcriptional activity (coding and non-coding) can help you get a deeper understanding of biology using NGS-based RNA-Seq.
We offer technologies to go beyond traditional cell and molecular biology research techniques so you can harness the power of genomic, transcriptomic, and epigenomic data efficiently and accurately.
FFPE RNA-Seq from degraded tissues
Get high-quality results from formalin-fixed, paraffin-embedded (FFPE) tissues using Illumina RNA-Seq solutions.
Your methods guide for NGS-based applications in cancer research, including spatial transcriptomics, is available now.
Gene expression and regulation research guide
See how Illumina RNA-Seq solutions can empower transcriptomic and epigenetic methods to advance discoveries in biology.
Your email address is never shared with third parties.
Want to learn more about spatial transcriptomics?
Your email address is never shared with third parties.