
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
The first CE-marked IVD kitted solution for comprehensive genomic profiling (CGP) of DNA and RNA variants, plus MSI and TMB, for multiple solid tumor types.
This product is only available in select locations. Please select the location where you would like this product to be shipped to see availability.
TruSight Oncology Comprehensive (EU) is a CE-marked IVD next-generation sequencing (NGS)-based CGP for analyzing > 28 solid tumor types using minimal tissue.
Content includes key biomarkers associated with guidelines, drug labels, European Society for Medical Oncology (ESMO) recommendations, and clinical trials.
TruSight Oncology Comprehensive (EU) is indicated as a companion diagnostic (CDx) test to identify cancer patients with solid tumors who are positive for NTRK1, NTRK2, or NTRK3 gene fusions, for treatment with VITRAKVI (larotrectinib) in accordance with the approved therapeutic labeling.
Additional companion diagnostic claims are under development.
Rely on a distributed sample-to-answer solution that can be implemented by local labs. Offer precision oncology in your institution and keep the data and sample in-house, reducing the likelihood of quantity not sufficient (QNS) issues.
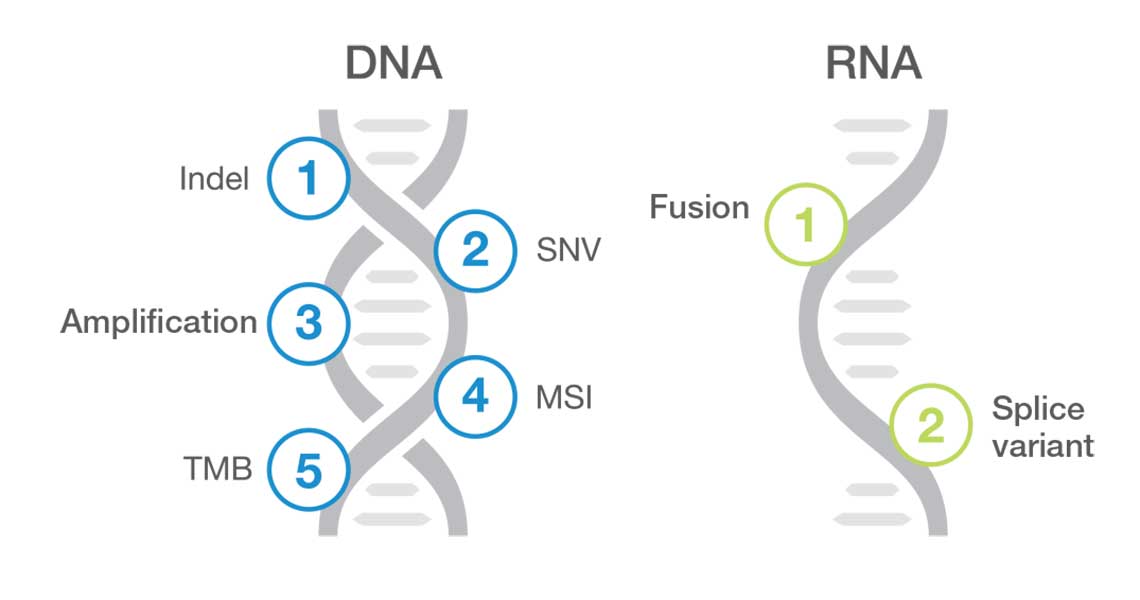
TruSight Oncology Comprehensive (EU) is an in vitro diagnostic test that uses targeted next generation sequencing to detect variants in 517 genes using nucleic acids extracted from formalin-fixed, paraffin embedded (FFPE) tumor tissue samples from cancer patients with solid malignant neoplasms using the Illumina NextSeq 550Dx instrument. The test can be used to detect single nucleotide variants, multinucleotide variants, insertions, deletions and gene amplifications from DNA, and gene fusions and splice variants from RNA. The test also reports a Tumor Mutational Burden (TMB) score and Microsatellite Instability (MSI) status.
The test is intended as a companion diagnostic to identify cancer patients for treatment with the targeted therapy listed in Table 1, in accordance with the approved therapeutic product labeling. In addition, the test is intended to provide tumor profiling information for use by qualified healthcare professionals in accordance with professional guidelines and is not conclusive or prescriptive for labeled use of any specific therapeutic product.
Table 1: Companion diagnostics indication
| Tumor Type | Biomarkers | Targeted Therapy |
|---|---|---|
| Solid Tumors | NTRK1, NTRK2, and NTRK3 gene fusions | VITRAKVI® (larotrectinib) |
| Assay time | 4-5 days |
|---|---|
| Content specifications | Covers clinically relevant biomarkers across 28+ solid tumors included in 64 clinical guidelines, 111 drug labels, and 615+ clinical trials. Panel content: 517 genes for small variants, 23 genes for fusions, 2 genes for splice variants (MET, EGFR), 2 genes for amplifications (ERBB2, MET), TMB and MSI. |
| False positive rate by rna variant type |
|
| False positives by dna variant type |
|
| Hands-on time | ~10.5 hr |
| Input quantity | 40 ng genomic DNA and 40 ng total RNA |
| Instruments | NextSeq 550Dx Instrument |
| Method | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment |
| Nucleic acid type | DNA, RNA |
| Sample throughput | Up to 7 patients and 2 controls (1 positive and 1 NTC) samples per sequencing run |
| Species category | Human |
| Technology | Sequencing |
TruSight Oncology Comprehensive (EU) is an in vitro diagnostic test that generates a comprehensive genomic profile of a patient’s tumor, informing therapy decisions according to clinical guidelines.
TruSight Oncology Comprehensive (EU)
Local Run Manager TruSight Oncology Comprehensive (EU) Analysis Module (on-instrument software)
Comprehensive genomic profiling
This next-generation sequencing approach consolidates hundreds of cancer-related markers, including different variant types, into a single assay.
Illumina NGS and microarray technologies for cancer research are helping drive the revolution in cancer genomics.


Batch up to seven patient samples and two control samples per run. Library prep and enrichment take 2 days, followed by a fully automated workflow on the NextSeq 550Dx Instrument. The entire workflow takes 4–5 days.



Reported variants categorized as clinically significant or potentially clinically significant based on an expertly curated knowledge base including clinical guidelines, drug labels, clinical trials, and peer-reviewed literature. Easy-to-read output helps increase confidence in treatment decisions.
| Tumor type | Selected genes with biomarkers of clinical significancea | |||||||
|---|---|---|---|---|---|---|---|---|
| BRAF | FGFR1 | FGFR2 | FGFR3 | MSI | NTRK1 | |||
| NTRK2 | NTRK3 | RET | TMB | |||||
| AKT1 | BRCA1 | BRCA2 | ERBB2 | ESR1 | PALB2 | |||
| PIK3CA | PTEN | |||||||
| BRAF | ERBB2 | KRAS | NRAS | POLE | MSI | |||
| BRAF | KIT | NRAS | ||||||
| ALK | BRAF | EGFR | ERBB2 | KRAS | MET | |||
| NRG1 | RET | ROS1 | ||||||
| BRCA1 | BRCA2 | |||||||
| BRCA1 | BRCA2 | KRAS | NRG1 | PALB2 | ||||
| ATM | BRCA1 | BRCA2 | PALB2 | PTEN | ||||
a. Genes with biomarkers of clinical significance linked to major oncology guidelines. MSI, microsatellite instability; TMB, tumor mutational burden.
TruSight Oncology Comprehensive (EU) Kit
20063092
Includes reagents for extraction, library preparation, and quantitation of 24 DNA and 24 RNA samples isolated from FFPE. Purchase NextSeq 550Dx sequencing reagents separately.
List Price:
Discounts:
TruSight Oncology DNA Control
20065041
Includes a qualitative IVD control for monitoring analytical performance of library prep, sequencing, and analysis.
List Price:
Discounts:
TruSight Oncology RNA Control
20065042
Includes a qualitative IVD control for monitoring analytical performance of library prep, sequencing, and analysis.
List Price:
Discounts:
NextSeq 550Dx High Output Reagent Kit v2.5 (300 cycles) IVD
20028871
NextSeq 550Dx High Output Reagent Kit v2.5 (300 Cycles) is a set of reagents and consumables intended for sequencing of sample libraries when used with validated assays. The kit is intended for use with the NextSeq 550Dx instrument and analytical software.
List Price:
Discounts:
NextSeq™ 550Dx Sequencing System
20005715
The NextSeq 550Dx instrument is intended for sequencing of DNA libraries when used with in vitro diagnostic assays performed on the instrument. The NextSeq 550Dx instrument is to be used with specific registered, certified or approved in vitro diagnostic reagents and analytical software.The instrument includes a dual boot configuration to enable the use of the instrument in either diagnostic (Dx) or research use only (RUO) mode. In vitro diagnostic sequencing assays, including the Germline and Somatic Variant Modules, are executed in diagnostic mode. Only IVD sequencing reagents can be utilized in diagnostic mode.
TSO Comprehensive Enablement Services
20066472
One-time required purchase that includes four days of hands-on training and workflow instruction (library prep, enrichment, sequencing, and data analysis) for up to two operators.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
The test was validated across > 350 unique FFPE samples and > 55 different tumor types. Results were compared to orthogonal methods to ensure accurate, reproducible, and consistent results.
Illumina offers a comprehensive support program that provides onboarding to expedite test verification, lab training, verification protocols, training certification, 24/5 technical support, support from our Medical Affairs team for medical inquiries, and educational and marketing assets to share with your local health care providers. Contact your local Illumina Account Manager for more details about the comprehensive support program.
Reimbursement differs based on the country, clinical setting, and services provided. Currently, national or regional funding is available in some European countries. Illumina has established a dedicated Market Access team that is actively working with payers to further expand CGP test reimbursement across the globe. Contact your local Illumina Account Manager with questions about coverage.
Illumina has established multiple partnerships with pharma companies to develop a growing pipeline of CDx indications pending regulatory approvals, that include but are not limited to RET (Eli Lilly),1 ROS1 (Roche),2 HRD (Myriad Genetics, Merck),3,4 and MSI (Bristol Myers Squibb).3
The minimum recommended tissue volume is 1 mm3 with a minimum of 20% tumor cell content by area required to detect somatic driver mutations; ≥ 30% tumor content is required to detect MSI-high. A minimum of five biopsy slides is recommended (10 µM sections, 20 mm2 tissue area each).

Contact us today.
Your email address is never shared with third parties.
References
1. lllumina and Loxo Oncology to partner on developing next-generation sequencing-based pan-cancer companion diagnostics. https://www.businesswire.com/news/home/20180410005649/en/. Accessed July 24, 2023.
2. Roche, Illumina partner on next-generation sequencing IVD, CDx development, marketing. https://www. genomeweb.com/business-news/roche-illumina-partnernext-generation-sequencing-ivd-cdx-developmentmarketing#.YWhVkhrMKUk. Accessed July 24, 2023.
3. Illumina announces new and expanded oncology partnerships with Bristol Myers Squibb, Kura Oncology, Myriad Genetics, and Merck to advance comprehensive genomic profiling. https://www.businesswire.com/news/home/20210111005930/en/Illumina-Announces-New-and-Expanded-Oncology-Partnerships-with-Bristol-Myers-Squibb-Kura-Oncology-Myriad-Genetics-and-Merck-to-Advance-Comprehensive-Genomic-Profiling. Accessed July 24, 2023.
4. Illumina partners with Merck to develop and commercialize companion diagnostic and research tests for use in identifying specific cancer mutations. https://www.prnewswire.com/news-releases/illumina-partners-with-merck-to-develop-and-commercialize-companion-diagnostic-and-research-tests-for-use-in-identifying-specific-cancer-mutations-301369838.html. Accessed July 24, 2023.
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.